Tutorial#
In this tutorial we demonstrate the use of the CoRegTor tool to find transcription co regulators for a gene from gene expression data.
Objective#
The aim of this tutorial is to find potential co-regulators of the gene GFAP by analyzing tissue gene expression data for Frontal cortex in adult brain.
Step 1 : Install and import the CoRegTor package and other dependencies#
Using pip, pip install coregtor .
Or poetry install coregtor to add the package as a dependency in your project
# Install coregtor if not already installed, then import it
try:
import coregtor
except ImportError:
%pip install coregtor
import coregtor
# Additional imports
from pathlib import Path
import pandas as pd
Step 2 : Get data and load it#
Let’s gather all the data we require:
Gene Expression data
ge_brain.gct. This file contains tissue gene expression data for the Frontal Cortex (BA9) in an adult brain. The data is downloaded from the GTEx portalList of transcription factors
human_tf.txt: This file was downloaded from aertslab.org
We load this data in dataframes. Now we are ready to use CoRegTor!
base_path = Path("docs/temp") # UPDATE THIS
data_file_path = Path(base_path/"brain_ge.gct") # UPDATE THIS
tf_file_path = Path(base_path/"human_tf.txt") # UPDATE THIS
target_gene_name = "GFAP" # the gene we are interested in
# load data
ge_data = coregtor.utils.read_GE_data(file_path=data_file_path) # this is just a utility method
tf_data = pd.read_csv(tf_file_path, names=["gene_name"], header=None)
ge_data
| gene_name | DDX11L1 | WASH7P | MIR6859-1 | MIR1302-2HG | FAM138A | OR4G4P | OR4G11P | OR4F5 | ENSG00000238009 | CICP27 | ... | MT-ND4 | MT-TH | MT-TS2 | MT-TL2 | MT-ND5 | MT-ND6 | MT-TE | MT-CYB | MT-TT | MT-TP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sample_name | |||||||||||||||||||||
| GTEX-1117F-0011-R10b-SM-GI4VE | 0.000000 | 3.57928 | 0.0 | 0.093825 | 0.000000 | 0.000000 | 0.028731 | 0.046554 | 0.039501 | 0.058675 | ... | 49762.2 | 1.177570 | 2.754330 | 0.000000 | 7311.39 | 4788.56 | 6.47666 | 28676.5 | 3.077750 | 1.19489 |
| GTEX-111FC-0011-R10a-SM-GIN8G | 0.000000 | 2.32926 | 0.0 | 0.025333 | 0.000000 | 0.052233 | 0.031030 | 0.016759 | 0.000000 | 0.031684 | ... | 44692.0 | 0.953824 | 0.000000 | 1.544930 | 6831.00 | 5164.36 | 6.67677 | 26950.9 | 1.661970 | 3.54879 |
| GTEX-117XS-0011-R10b-SM-GIN8Z | 0.000000 | 4.79425 | 0.0 | 0.000000 | 0.046843 | 0.067977 | 0.020191 | 0.043622 | 0.013880 | 0.032987 | ... | 39249.9 | 0.827551 | 0.967814 | 1.206360 | 5603.53 | 3585.51 | 6.20663 | 20794.9 | 0.432584 | 2.93902 |
| GTEX-1192W-0011-R10b-SM-GHWOF | 0.000000 | 3.83774 | 0.0 | 0.032159 | 0.045693 | 0.000000 | 0.039392 | 0.053189 | 0.013539 | 0.000000 | ... | 50750.5 | 1.614480 | 2.832190 | 1.176750 | 9433.33 | 7697.90 | 12.51220 | 23405.4 | 1.265900 | 3.68601 |
| GTEX-1192X-0011-R10a-SM-DO941 | 0.040388 | 1.47233 | 0.0 | 0.040318 | 0.000000 | 0.000000 | 0.049385 | 0.040010 | 0.050922 | 0.000000 | ... | 31566.9 | 2.024070 | 0.591784 | 0.983528 | 4424.64 | 3568.41 | 4.55416 | 14051.5 | 0.529019 | 1.54038 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| GTEX-ZVZQ-0011-R10b-SM-51MRT | 0.017553 | 1.91964 | 0.0 | 0.070089 | 0.000000 | 0.180647 | 0.064389 | 0.092738 | 0.044262 | 0.043831 | ... | 44939.6 | 3.078850 | 1.543150 | 2.137230 | 7019.94 | 6874.29 | 16.71380 | 24296.2 | 1.379490 | 2.23152 |
| GTEX-ZXG5-0011-R10a-SM-57WDD | 0.000000 | 1.07536 | 0.0 | 0.036646 | 0.000000 | 0.000000 | 0.044887 | 0.084853 | 0.000000 | 0.000000 | ... | 62226.7 | 2.759570 | 1.613650 | 4.469730 | 11407.90 | 11061.80 | 15.17770 | 38732.2 | 1.442500 | 1.86677 |
| GTEX-ZYFD-0011-R10a-SM-GPI91 | 0.000000 | 2.71020 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.037432 | 0.000000 | 0.000000 | 0.030577 | ... | 43740.3 | 0.000000 | 2.691290 | 0.745473 | 6574.39 | 5241.85 | 9.20498 | 24934.3 | 0.000000 | 1.55672 |
| GTEX-ZYY3-0011-R10a-SM-GNTAZ | 0.015919 | 3.29538 | 0.0 | 0.000000 | 0.000000 | 0.065533 | 0.058395 | 0.031540 | 0.066902 | 0.079502 | ... | 40835.8 | 1.196680 | 0.933006 | 3.101260 | 6228.45 | 5626.94 | 10.37120 | 20992.5 | 5.004310 | 2.83332 |
| GTEX-ZZPT-0011-R10b-SM-GPI8B | 0.000000 | 2.85899 | 0.0 | 0.049821 | 0.000000 | 0.000000 | 0.030513 | 0.000000 | 0.041949 | 0.024925 | ... | 40128.3 | 1.250570 | 0.000000 | 1.215350 | 8300.34 | 8785.10 | 28.13790 | 30362.8 | 2.614830 | 3.80689 |
269 rows × 59033 columns
tf_data
| gene_name | |
|---|---|
| 0 | ZNF354C |
| 1 | KLF12 |
| 2 | ZNF143 |
| 3 | ZIC2 |
| 4 | ZNF274 |
| ... | ... |
| 1887 | ZNF826P |
| 1888 | ZNF827 |
| 1889 | ZNF831 |
| 1890 | ZRSR2 |
| 1891 | ZSWIM1 |
1892 rows × 1 columns
Step 3 : Create Ensemble model#
The first step in the process is to generate a random forest ensemble model using the gene expression data that predicts the expression value of the gene “GFAP” based on all other genes.
Since we are interested in identifying potential transcription co regulators, we filter the data to include only transcription factors. We use the create_model_input function for filtering and preparing the input for training. It takes a dataframe t_factors which should have a column gene_name listing the transcription factors to consider. The function outputs a tuple containing 2 data frames: X with the feature genes and Y with the target gene. These can then be passed to the generate_model function.
# first generate the training input for the model
X,Y = coregtor.create_model_input(ge_data,target_gene_name,tf_data)
# use the training data to create a model
model = coregtor.create_model(X,Y,"rf")
model
RandomForestRegressor()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Parameters
| n_estimators | 100 | |
| criterion | 'squared_error' | |
| max_depth | None | |
| min_samples_split | 2 | |
| min_samples_leaf | 1 | |
| min_weight_fraction_leaf | 0.0 | |
| max_features | 1.0 | |
| max_leaf_nodes | None | |
| min_impurity_decrease | 0.0 | |
| bootstrap | True | |
| oob_score | False | |
| n_jobs | None | |
| random_state | None | |
| verbose | 0 | |
| warm_start | False | |
| ccp_alpha | 0.0 | |
| max_samples | None | |
| monotonic_cst | None |
Step 4 : Generating tree paths#
The genes on the root node are important. These also serve as potential regulators in other tree based GRN inference methods.
Forest based ensemble methods contains multiple decision tress. We want to analyze the structure of the trees in the model.
For each tree, there exists multiple root to leaf paths. We first enumerate all the paths in all the trees in the model.
all_paths = coregtor.tree_paths(model,X,Y)
all_paths
| tree | source | target | path_length | node1 | node2 | node3 | node4 | node5 | node6 | ... | node9 | node10 | node11 | node12 | node13 | node14 | node15 | node16 | node17 | node18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | YBX1 | GFAP | 8 | TEAD2 | HES7 | ISL1 | LHX4 | ZNF135 | ZNF536 | ... | None | None | None | None | None | None | None | None | None | None |
| 1 | 0 | YBX1 | GFAP | 12 | TEAD2 | TSC22D4 | PAX5 | EBF1 | GSX2 | TFEB | ... | VENTX | PAX3 | ZNF70 | None | None | None | None | None | None | None |
| 2 | 0 | YBX1 | GFAP | 13 | TEAD2 | TSC22D4 | PAX5 | EBF1 | GSX2 | TFEB | ... | VENTX | PAX3 | NR1I2 | TBX21 | None | None | None | None | None | None |
| 3 | 0 | YBX1 | GFAP | 6 | POU5F1B | ZNF433 | GLIS3 | ZKSCAN3 | ZNF436 | None | ... | None | None | None | None | None | None | None | None | None | None |
| 4 | 0 | YBX1 | GFAP | 10 | TEAD2 | TSC22D4 | PAX5 | SNAI2 | RFX6 | HOXA5 | ... | SMAD2 | None | None | None | None | None | None | None | None | None |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 11013 | 99 | FEZ1 | GFAP | 8 | YBX1 | IRF9 | HMG20B | ZBTB18 | ZNF683 | ZNF556 | ... | None | None | None | None | None | None | None | None | None | None |
| 11014 | 99 | FEZ1 | GFAP | 14 | YBX1 | IRF9 | HMG20B | ZBTB18 | TFEB | NKX2-2 | ... | ZNF683 | TBX5 | BHLHE40 | MAGEA8 | E2F3 | None | None | None | None | None |
| 11015 | 99 | FEZ1 | GFAP | 15 | YBX1 | IRF9 | HMG20B | ZBTB18 | TFEB | NKX2-2 | ... | TBX22 | FOXP1 | CREB3L4 | ZNF554 | MAP4K2 | ZNF814 | None | None | None | None |
| 11016 | 99 | FEZ1 | GFAP | 14 | YBX1 | IRF9 | HMG20B | ZBTB18 | TFEB | NKX2-2 | ... | TBX22 | FOXP1 | CREB3L4 | ZNF554 | MAP4K2 | None | None | None | None | None |
| 11017 | 99 | FEZ1 | GFAP | 7 | YBX1 | IRF9 | HMG20B | TAF1L | NHLH1 | NAP1L1 | ... | None | None | None | None | None | None | None | None | None | None |
11018 rows × 22 columns
Step 5 : Generating a set of common sub paths (or the context) for all root nodes#
In the table of paths above, we observe many unique genes appear as root nodes. Since we train the decision trees to predict the same target gene, the leaf nodes for all paths are the same.
We can thus consider these paths as potential regulatory links, where genes at the root regulates the target. All the genes at the root become potential regulators or the target. However, note that there are multiple intermediate nodes between the root and the target. Comparing these
We consider the root nodes as potential regulators of the target gene and to find if they are co regulators, we compare how similar the intermediate nodes are in between 2 unique root nodes.
pathset = coregtor.create_context(all_paths)
pathset.keys()
dict_keys(['YBX1', 'POU2AF1', 'PIR', 'ALX3', 'HCLS1', 'SP110', 'TIGD1', 'ZNF577', 'TAGLN2', 'KLF15', 'ZRSR2', 'FOXB2', 'ZNF837', 'IRF9', 'HMBOX1', 'PPP2R3B', 'DPRX', 'NFYA', 'ZNF775', 'SMC3', 'ID1', 'ZNF768', 'NKX6-2', 'ZNF706', 'CLK1', 'HDAC1', 'ZNF322', 'RAD21', 'PHF21A', 'MAFK', 'CREB3L2', 'TBX19', 'HTATIP2', 'ZSCAN16', 'IKZF2', 'SOX10', 'MXI1', 'BCL6', 'FEZ1'])
Step 6 : Comparing context of all root nodes with each other#
similarity
# transforming the context into a more comparable representation
gf_histogram = coregtor.transform_context(pathset,method="gene_frequency")
gf_histogram
| HES7 | ISL1 | LHX4 | ZNF135 | ZNF536 | TSC22D4 | PAX5 | EBF1 | GSX2 | TFEB | ... | ZNF34 | TCEAL6 | RXRB | FOXI1 | PSMD12 | KLF12 | ZNF783 | ZNF749 | ZCCHC14 | FOXP1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YBX1 | 8 | 4 | 4 | 3 | 4 | 362 | 143 | 42 | 42 | 26 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| POU2AF1 | 0 | 5 | 0 | 0 | 0 | 74 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PIR | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ALX3 | 2 | 0 | 0 | 0 | 0 | 61 | 0 | 0 | 0 | 2 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HCLS1 | 0 | 0 | 1 | 0 | 0 | 63 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SP110 | 0 | 0 | 0 | 0 | 0 | 48 | 47 | 6 | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TIGD1 | 0 | 0 | 0 | 0 | 0 | 72 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ZNF577 | 0 | 0 | 0 | 0 | 0 | 3 | 21 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TAGLN2 | 0 | 0 | 3 | 0 | 0 | 69 | 12 | 0 | 5 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| KLF15 | 1 | 0 | 0 | 0 | 0 | 139 | 22 | 0 | 0 | 6 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ZRSR2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FOXB2 | 0 | 0 | 0 | 0 | 1 | 178 | 1 | 14 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ZNF837 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IRF9 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HMBOX1 | 3 | 2 | 0 | 0 | 12 | 443 | 117 | 0 | 11 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PPP2R3B | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 6 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| DPRX | 0 | 0 | 1 | 0 | 1 | 114 | 10 | 1 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NFYA | 0 | 0 | 0 | 0 | 0 | 88 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ZNF775 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 4 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SMC3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ID1 | 0 | 0 | 0 | 0 | 0 | 57 | 0 | 3 | 30 | 5 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ZNF768 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NKX6-2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ZNF706 | 0 | 1 | 0 | 0 | 2 | 33 | 0 | 0 | 7 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CLK1 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HDAC1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ZNF322 | 0 | 0 | 0 | 0 | 0 | 66 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RAD21 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PHF21A | 0 | 0 | 2 | 0 | 0 | 79 | 0 | 1 | 5 | 0 | ... | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| MAFK | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CREB3L2 | 0 | 0 | 0 | 0 | 0 | 44 | 42 | 0 | 2 | 0 | ... | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TBX19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | ... | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| HTATIP2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | ... | 0 | 0 | 0 | 0 | 1 | 4 | 1 | 0 | 0 | 0 |
| ZSCAN16 | 0 | 0 | 0 | 0 | 0 | 45 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IKZF2 | 0 | 0 | 0 | 0 | 0 | 30 | 14 | 0 | 0 | 3 | ... | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| SOX10 | 0 | 0 | 0 | 0 | 0 | 32 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| MXI1 | 2 | 0 | 0 | 0 | 0 | 0 | 17 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| BCL6 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FEZ1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
39 rows × 1815 columns
sim_matrix = coregtor.compare_context(gf_histogram,"cosine")
sim_matrix
| YBX1 | POU2AF1 | PIR | ALX3 | HCLS1 | SP110 | TIGD1 | ZNF577 | TAGLN2 | KLF15 | ... | MAFK | CREB3L2 | TBX19 | HTATIP2 | ZSCAN16 | IKZF2 | SOX10 | MXI1 | BCL6 | FEZ1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YBX1 | 1.000000 | 0.390690 | 0.282963 | 0.441397 | 0.347206 | 0.357401 | 0.437412 | 0.178489 | 0.427796 | 0.527324 | ... | 0.207780 | 0.443300 | 0.268885 | 0.110692 | 0.464645 | 0.458956 | 0.444353 | 0.229340 | 0.132132 | 0.048082 |
| POU2AF1 | 0.390690 | 1.000000 | 0.097367 | 0.189427 | 0.191951 | 0.271433 | 0.280371 | 0.039441 | 0.151647 | 0.246245 | ... | 0.057232 | 0.274348 | 0.135393 | 0.009952 | 0.208366 | 0.147100 | 0.172599 | 0.025864 | 0.054748 | 0.272656 |
| PIR | 0.282963 | 0.097367 | 1.000000 | 0.271940 | 0.081994 | 0.174818 | 0.071998 | 0.030674 | 0.101351 | 0.290197 | ... | 0.055558 | 0.295340 | 0.404131 | 0.010030 | 0.387235 | 0.326790 | 0.390717 | 0.013391 | 0.030086 | 0.006771 |
| ALX3 | 0.441397 | 0.189427 | 0.271940 | 1.000000 | 0.462176 | 0.183100 | 0.205235 | 0.306286 | 0.466232 | 0.456986 | ... | 0.297251 | 0.291815 | 0.278464 | 0.088860 | 0.355313 | 0.272073 | 0.332201 | 0.017341 | 0.036216 | 0.077981 |
| HCLS1 | 0.347206 | 0.191951 | 0.081994 | 0.462176 | 1.000000 | 0.182966 | 0.269786 | 0.328728 | 0.485007 | 0.328482 | ... | 0.336335 | 0.152378 | 0.052297 | 0.017688 | 0.166456 | 0.104280 | 0.114887 | 0.083077 | 0.110200 | 0.024756 |
| SP110 | 0.357401 | 0.271433 | 0.174818 | 0.183100 | 0.182966 | 1.000000 | 0.251293 | 0.045317 | 0.140399 | 0.264442 | ... | 0.066215 | 0.278668 | 0.182943 | 0.060086 | 0.228899 | 0.191671 | 0.188913 | 0.135039 | 0.049885 | 0.072055 |
| TIGD1 | 0.437412 | 0.280371 | 0.071998 | 0.205235 | 0.269786 | 0.251293 | 1.000000 | 0.027062 | 0.219067 | 0.350281 | ... | 0.023705 | 0.221418 | 0.030539 | 0.049867 | 0.235625 | 0.214205 | 0.204016 | 0.124777 | 0.068017 | 0.005000 |
| ZNF577 | 0.178489 | 0.039441 | 0.030674 | 0.306286 | 0.328728 | 0.045317 | 0.027062 | 1.000000 | 0.344919 | 0.170948 | ... | 0.279431 | 0.082814 | 0.004852 | 0.069558 | 0.039215 | 0.026043 | 0.008941 | 0.027977 | 0.024586 | 0.001571 |
| TAGLN2 | 0.427796 | 0.151647 | 0.101351 | 0.466232 | 0.485007 | 0.140399 | 0.219067 | 0.344919 | 1.000000 | 0.309809 | ... | 0.433702 | 0.177477 | 0.062660 | 0.066611 | 0.173707 | 0.171206 | 0.169182 | 0.281993 | 0.025474 | 0.040933 |
| KLF15 | 0.527324 | 0.246245 | 0.290197 | 0.456986 | 0.328482 | 0.264442 | 0.350281 | 0.170948 | 0.309809 | 1.000000 | ... | 0.132012 | 0.342737 | 0.285807 | 0.428690 | 0.413125 | 0.599421 | 0.434714 | 0.308292 | 0.299045 | 0.192649 |
| ZRSR2 | 0.033044 | 0.263826 | 0.008146 | 0.132157 | 0.014655 | 0.058607 | 0.010481 | 0.027319 | 0.078656 | 0.047919 | ... | 0.022265 | 0.110977 | 0.006141 | 0.004314 | 0.005255 | 0.011769 | 0.017606 | 0.006950 | 0.028671 | 0.314943 |
| FOXB2 | 0.610984 | 0.357531 | 0.276302 | 0.345941 | 0.287703 | 0.249759 | 0.464244 | 0.024721 | 0.279255 | 0.550246 | ... | 0.014182 | 0.369243 | 0.254869 | 0.171139 | 0.487059 | 0.503505 | 0.461591 | 0.202323 | 0.219223 | 0.006372 |
| ZNF837 | 0.295305 | 0.064716 | 0.330254 | 0.247707 | 0.039826 | 0.184614 | 0.047311 | 0.007792 | 0.188687 | 0.248245 | ... | 0.072768 | 0.236638 | 0.390586 | 0.004310 | 0.359435 | 0.313760 | 0.357238 | 0.156812 | 0.016106 | 0.002320 |
| IRF9 | 0.295182 | 0.064347 | 0.335614 | 0.376965 | 0.061234 | 0.166046 | 0.039170 | 0.009453 | 0.094527 | 0.300712 | ... | 0.005489 | 0.259299 | 0.464632 | 0.122566 | 0.338762 | 0.285473 | 0.344640 | 0.028046 | 0.042869 | 0.098552 |
| HMBOX1 | 0.647789 | 0.329978 | 0.241217 | 0.462533 | 0.423149 | 0.397572 | 0.376354 | 0.216785 | 0.415346 | 0.647799 | ... | 0.189737 | 0.395000 | 0.263515 | 0.341196 | 0.382428 | 0.520478 | 0.392212 | 0.374563 | 0.368752 | 0.215369 |
| PPP2R3B | 0.048033 | 0.009664 | 0.016664 | 0.039575 | 0.145264 | 0.053923 | 0.015605 | 0.030973 | 0.032809 | 0.009607 | ... | 0.006765 | 0.034600 | 0.139428 | 0.054337 | 0.041394 | 0.004065 | 0.010393 | 0.018624 | 0.011406 | 0.044133 |
| DPRX | 0.465090 | 0.272496 | 0.073816 | 0.339412 | 0.450710 | 0.202101 | 0.390178 | 0.181192 | 0.475072 | 0.414972 | ... | 0.234993 | 0.192898 | 0.051587 | 0.114427 | 0.204696 | 0.246931 | 0.198077 | 0.253205 | 0.199409 | 0.049899 |
| NFYA | 0.438038 | 0.296309 | 0.086302 | 0.221245 | 0.392647 | 0.187856 | 0.436239 | 0.018539 | 0.211032 | 0.328365 | ... | 0.005740 | 0.246735 | 0.087293 | 0.015994 | 0.331019 | 0.177932 | 0.235494 | 0.012583 | 0.001819 | 0.030263 |
| ZNF775 | 0.288816 | 0.076733 | 0.302994 | 0.202763 | 0.010507 | 0.131376 | 0.064825 | 0.004195 | 0.105839 | 0.472321 | ... | 0.023656 | 0.245449 | 0.344232 | 0.332860 | 0.315589 | 0.621905 | 0.406507 | 0.399331 | 0.325837 | 0.000000 |
| SMC3 | 0.274880 | 0.362013 | 0.310281 | 0.251701 | 0.012456 | 0.184213 | 0.003516 | 0.057674 | 0.071365 | 0.228840 | ... | 0.061498 | 0.354918 | 0.384980 | 0.009969 | 0.325361 | 0.273689 | 0.324689 | 0.007600 | 0.019026 | 0.349449 |
| ID1 | 0.556863 | 0.264554 | 0.300524 | 0.303235 | 0.211422 | 0.221503 | 0.240994 | 0.023985 | 0.303998 | 0.486265 | ... | 0.128613 | 0.294769 | 0.379898 | 0.229686 | 0.394576 | 0.519505 | 0.446451 | 0.403588 | 0.210834 | 0.061608 |
| ZNF768 | 0.272612 | 0.056057 | 0.376421 | 0.261556 | 0.033323 | 0.198101 | 0.020632 | 0.040113 | 0.069010 | 0.298104 | ... | 0.018190 | 0.282073 | 0.441665 | 0.017832 | 0.386520 | 0.346977 | 0.395583 | 0.014390 | 0.202221 | 0.067571 |
| NKX6-2 | 0.285358 | 0.062473 | 0.415863 | 0.284157 | 0.021398 | 0.202981 | 0.017929 | 0.002389 | 0.077482 | 0.285184 | ... | 0.047676 | 0.297630 | 0.465344 | 0.017281 | 0.452095 | 0.359295 | 0.428401 | 0.023911 | 0.020326 | 0.002228 |
| ZNF706 | 0.359139 | 0.291627 | 0.266523 | 0.466796 | 0.329536 | 0.240143 | 0.139865 | 0.229249 | 0.335848 | 0.412629 | ... | 0.240955 | 0.277752 | 0.377293 | 0.048290 | 0.323392 | 0.305714 | 0.374505 | 0.069505 | 0.136062 | 0.114209 |
| CLK1 | 0.247502 | 0.070450 | 0.353188 | 0.384848 | 0.004387 | 0.157336 | 0.005762 | 0.018712 | 0.156468 | 0.305405 | ... | 0.003527 | 0.256807 | 0.413187 | 0.130467 | 0.377501 | 0.318310 | 0.376075 | 0.004852 | 0.003137 | 0.130224 |
| HDAC1 | 0.065214 | 0.015740 | 0.021484 | 0.021804 | 0.280865 | 0.070350 | 0.016731 | 0.080937 | 0.196380 | 0.007186 | ... | 0.006690 | 0.048311 | 0.012432 | 0.081104 | 0.043150 | 0.010721 | 0.034793 | 0.148792 | 0.129979 | 0.008133 |
| ZNF322 | 0.383818 | 0.326568 | 0.063726 | 0.226584 | 0.234236 | 0.161013 | 0.374197 | 0.013847 | 0.184562 | 0.257207 | ... | 0.009382 | 0.228177 | 0.014658 | 0.000900 | 0.237554 | 0.151398 | 0.189613 | 0.009085 | 0.002382 | 0.105703 |
| RAD21 | 0.255462 | 0.067092 | 0.339623 | 0.269857 | 0.013692 | 0.224968 | 0.002939 | 0.013755 | 0.077211 | 0.284662 | ... | 0.001262 | 0.270840 | 0.416535 | 0.014091 | 0.355123 | 0.365356 | 0.399475 | 0.011585 | 0.003843 | 0.009626 |
| PHF21A | 0.442503 | 0.189444 | 0.217075 | 0.618650 | 0.599663 | 0.183091 | 0.260008 | 0.401262 | 0.497952 | 0.569636 | ... | 0.334362 | 0.246240 | 0.234853 | 0.193900 | 0.294922 | 0.417305 | 0.354604 | 0.199902 | 0.208485 | 0.042961 |
| MAFK | 0.207780 | 0.057232 | 0.055558 | 0.297251 | 0.336335 | 0.066215 | 0.023705 | 0.279431 | 0.433702 | 0.132012 | ... | 1.000000 | 0.008271 | 0.005600 | 0.008667 | 0.011967 | 0.016547 | 0.011851 | 0.163160 | 0.018630 | 0.024411 |
| CREB3L2 | 0.443300 | 0.274348 | 0.295340 | 0.291815 | 0.152378 | 0.278668 | 0.221418 | 0.082814 | 0.177477 | 0.342737 | ... | 0.008271 | 1.000000 | 0.269212 | 0.031385 | 0.393787 | 0.338274 | 0.341131 | 0.063796 | 0.016362 | 0.137304 |
| TBX19 | 0.268885 | 0.135393 | 0.404131 | 0.278464 | 0.052297 | 0.182943 | 0.030539 | 0.004852 | 0.062660 | 0.285807 | ... | 0.005600 | 0.269212 | 1.000000 | 0.007271 | 0.398668 | 0.350113 | 0.428055 | 0.022620 | 0.015436 | 0.015548 |
| HTATIP2 | 0.110692 | 0.009952 | 0.010030 | 0.088860 | 0.017688 | 0.060086 | 0.049867 | 0.069558 | 0.066611 | 0.428690 | ... | 0.008667 | 0.031385 | 0.007271 | 1.000000 | 0.033336 | 0.354398 | 0.088057 | 0.355422 | 0.302257 | 0.420583 |
| ZSCAN16 | 0.464645 | 0.208366 | 0.387235 | 0.355313 | 0.166456 | 0.228899 | 0.235625 | 0.039215 | 0.173707 | 0.413125 | ... | 0.011967 | 0.393787 | 0.398668 | 0.033336 | 1.000000 | 0.443097 | 0.476257 | 0.006599 | 0.010767 | 0.048056 |
| IKZF2 | 0.458956 | 0.147100 | 0.326790 | 0.272073 | 0.104280 | 0.191671 | 0.214205 | 0.026043 | 0.171206 | 0.599421 | ... | 0.016547 | 0.338274 | 0.350113 | 0.354398 | 0.443097 | 1.000000 | 0.479705 | 0.395397 | 0.316269 | 0.044545 |
| SOX10 | 0.444353 | 0.172599 | 0.390717 | 0.332201 | 0.114887 | 0.188913 | 0.204016 | 0.008941 | 0.169182 | 0.434714 | ... | 0.011851 | 0.341131 | 0.428055 | 0.088057 | 0.476257 | 0.479705 | 1.000000 | 0.123353 | 0.083377 | 0.008350 |
| MXI1 | 0.229340 | 0.025864 | 0.013391 | 0.017341 | 0.083077 | 0.135039 | 0.124777 | 0.027977 | 0.281993 | 0.308292 | ... | 0.163160 | 0.063796 | 0.022620 | 0.355422 | 0.006599 | 0.395397 | 0.123353 | 1.000000 | 0.338336 | 0.006669 |
| BCL6 | 0.132132 | 0.054748 | 0.030086 | 0.036216 | 0.110200 | 0.049885 | 0.068017 | 0.024586 | 0.025474 | 0.299045 | ... | 0.018630 | 0.016362 | 0.015436 | 0.302257 | 0.010767 | 0.316269 | 0.083377 | 0.338336 | 1.000000 | 0.070355 |
| FEZ1 | 0.048082 | 0.272656 | 0.006771 | 0.077981 | 0.024756 | 0.072055 | 0.005000 | 0.001571 | 0.040933 | 0.192649 | ... | 0.024411 | 0.137304 | 0.015548 | 0.420583 | 0.048056 | 0.044545 | 0.008350 | 0.006669 | 0.070355 | 1.000000 |
39 rows × 39 columns
Step 7 : Interactive generation of co-regulating gene clusters#
# Dendrogram
coregtor.plot_dendrogram(sim_matrix)
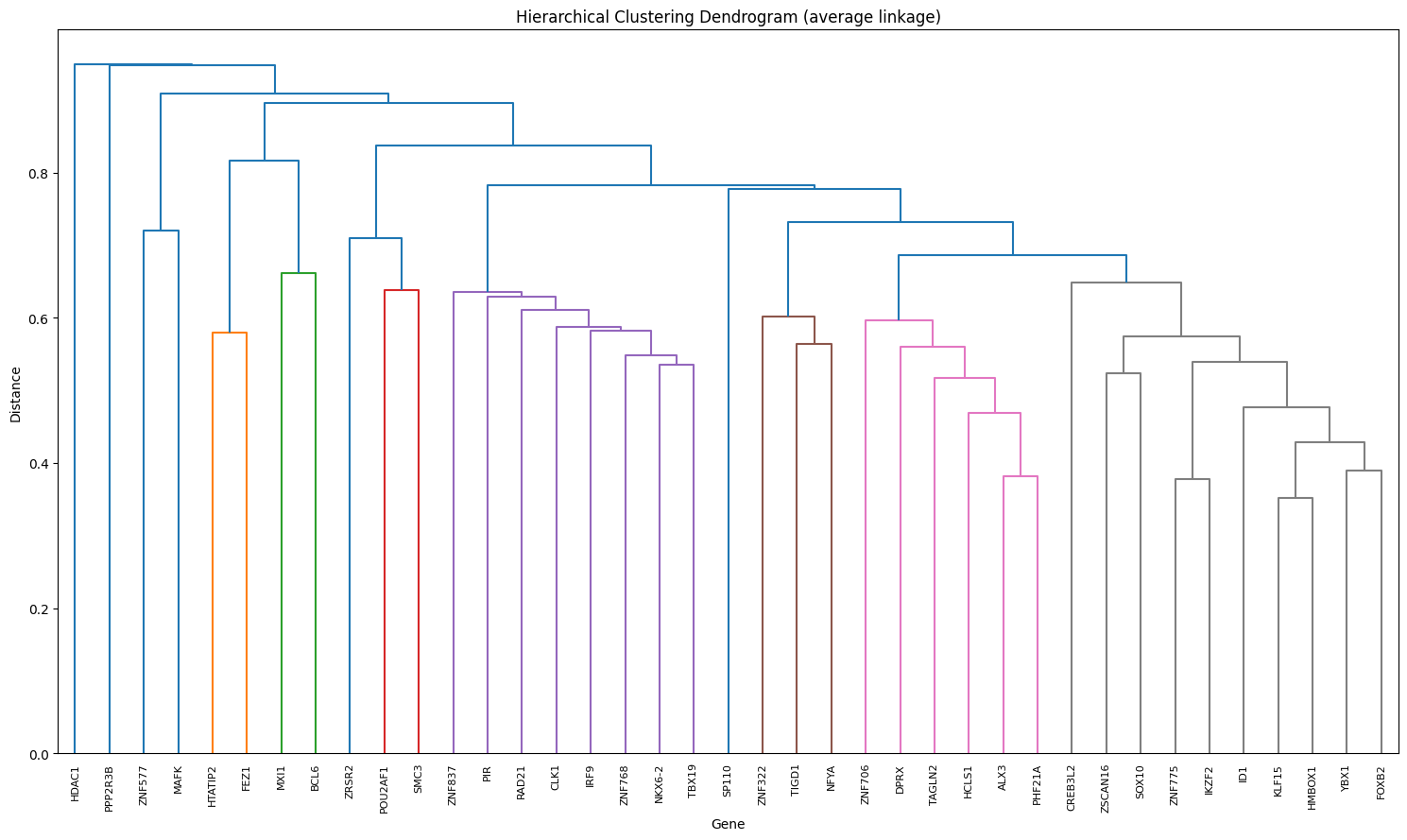
(<Figure size 1500x900 with 1 Axes>,
<Axes: title={'center': 'Hierarchical Clustering Dendrogram (average linkage)'}, xlabel='Gene', ylabel='Distance'>,
array([[ 9. , 14. , 0.35220059, 2. ],
[18. , 34. , 0.37809491, 2. ],
[ 3. , 28. , 0.38135024, 2. ],
[ 0. , 11. , 0.38901568, 2. ],
[39. , 42. , 0.42850833, 4. ],
[ 4. , 41. , 0.46908031, 3. ],
[20. , 43. , 0.47638252, 5. ],
[ 8. , 44. , 0.51693641, 4. ],
[33. , 35. , 0.52374254, 2. ],
[22. , 31. , 0.53465561, 2. ],
[40. , 45. , 0.53960328, 7. ],
[21. , 48. , 0.54866987, 3. ],
[16. , 46. , 0.56050718, 5. ],
[ 6. , 17. , 0.5637613 , 2. ],
[47. , 49. , 0.57385334, 9. ],
[32. , 38. , 0.57941738, 2. ],
[13. , 50. , 0.58256298, 4. ],
[24. , 55. , 0.58741151, 5. ],
[23. , 51. , 0.59591912, 6. ],
[26. , 52. , 0.60223391, 3. ],
[27. , 56. , 0.61093111, 6. ],
[ 2. , 59. , 0.62919343, 7. ],
[12. , 60. , 0.636039 , 8. ],
[ 1. , 19. , 0.63798693, 2. ],
[30. , 53. , 0.64847878, 10. ],
[36. , 37. , 0.661664 , 2. ],
[57. , 63. , 0.68634988, 16. ],
[10. , 62. , 0.70978445, 3. ],
[ 7. , 29. , 0.72056934, 2. ],
[58. , 65. , 0.73153913, 19. ],
[ 5. , 68. , 0.77672799, 20. ],
[61. , 69. , 0.78269649, 28. ],
[54. , 64. , 0.81632422, 4. ],
[66. , 70. , 0.83765542, 31. ],
[71. , 72. , 0.89525603, 35. ],
[67. , 73. , 0.90940586, 37. ],
[15. , 74. , 0.94759734, 38. ],
[25. , 75. , 0.94942835, 39. ]]))
# Cophonetic distance
coregtor.plot_cophenetic(sim_matrix,methods=["average"])
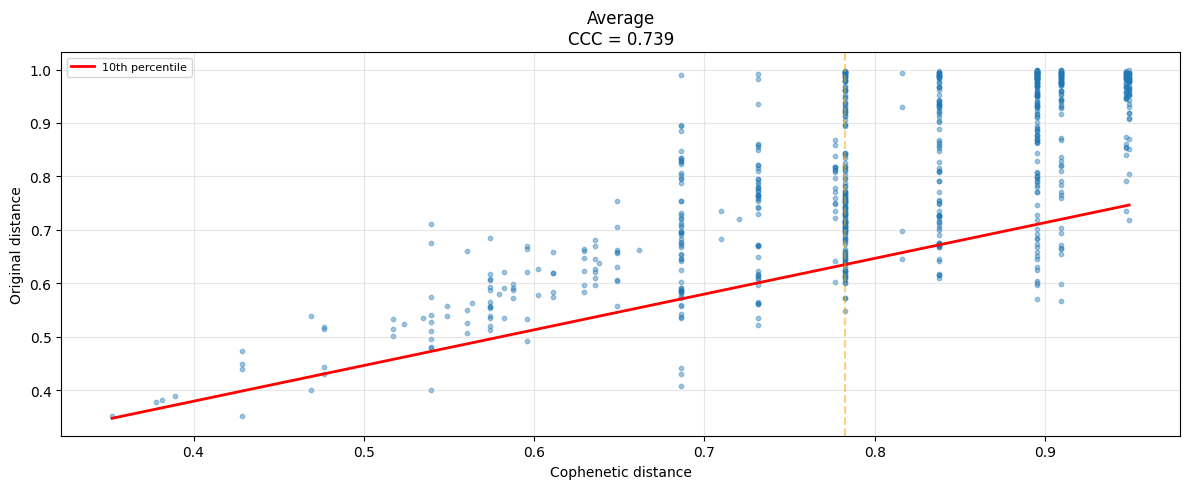
(<Figure size 1200x500 with 1 Axes>,
[<Axes: title={'center': 'Average\nCCC = 0.739'}, xlabel='Cophenetic distance', ylabel='Original distance'>],
{'average': np.float64(0.7391324338478736)},
np.float64(0.6350354525242414))
# generate results
results1,_ = coregtor.identify_coregulators(sim_matrix,target_gene=target_gene_name,distance_threshold=0.65)
results1
| target_gene | gene_cluster | n_genes | cluster_id | |
|---|---|---|---|---|
| 0 | GFAP | CREB3L2,FOXB2,HMBOX1,ID1,IKZF2,KLF15,SOX10,YBX... | 10 | 0 |
| 1 | GFAP | CLK1,IRF9,NKX6-2,PIR,RAD21,TBX19,ZNF768,ZNF837 | 8 | 3 |
| 2 | GFAP | ALX3,DPRX,HCLS1,PHF21A,TAGLN2,ZNF706 | 6 | 2 |
| 3 | GFAP | NFYA,TIGD1,ZNF322 | 3 | 4 |
| 4 | GFAP | POU2AF1,SMC3 | 2 | 1 |
| 5 | GFAP | FEZ1,HTATIP2 | 2 | 5 |
results1 = results1[results1["n_genes"]<10]
results1
| target_gene | gene_cluster | n_genes | cluster_id | |
|---|---|---|---|---|
| 1 | GFAP | CLK1,IRF9,NKX6-2,PIR,RAD21,TBX19,ZNF768,ZNF837 | 8 | 3 |
| 2 | GFAP | ALX3,DPRX,HCLS1,PHF21A,TAGLN2,ZNF706 | 6 | 2 |
| 3 | GFAP | NFYA,TIGD1,ZNF322 | 3 | 4 |
| 4 | GFAP | POU2AF1,SMC3 | 2 | 1 |
| 5 | GFAP | FEZ1,HTATIP2 | 2 | 5 |
results2,_ = coregtor.identify_coregulators(sim_matrix,target_gene=target_gene_name,distance_threshold=0.75)
results2
| target_gene | gene_cluster | n_genes | cluster_id | |
|---|---|---|---|---|
| 0 | GFAP | ALX3,CREB3L2,DPRX,FOXB2,HCLS1,HMBOX1,ID1,IKZF2... | 19 | 0 |
| 1 | GFAP | CLK1,IRF9,NKX6-2,PIR,RAD21,TBX19,ZNF768,ZNF837 | 8 | 3 |
| 2 | GFAP | POU2AF1,SMC3,ZRSR2 | 3 | 2 |
| 3 | GFAP | MAFK,ZNF577 | 2 | 1 |
| 4 | GFAP | BCL6,MXI1 | 2 | 4 |
| 5 | GFAP | FEZ1,HTATIP2 | 2 | 5 |
results2 = results2[results2["n_genes"]<10]
results2
| target_gene | gene_cluster | n_genes | cluster_id | |
|---|---|---|---|---|
| 1 | GFAP | CLK1,IRF9,NKX6-2,PIR,RAD21,TBX19,ZNF768,ZNF837 | 8 | 3 |
| 2 | GFAP | POU2AF1,SMC3,ZRSR2 | 3 | 2 |
| 3 | GFAP | MAFK,ZNF577 | 2 | 1 |
| 4 | GFAP | BCL6,MXI1 | 2 | 4 |
| 5 | GFAP | FEZ1,HTATIP2 | 2 | 5 |